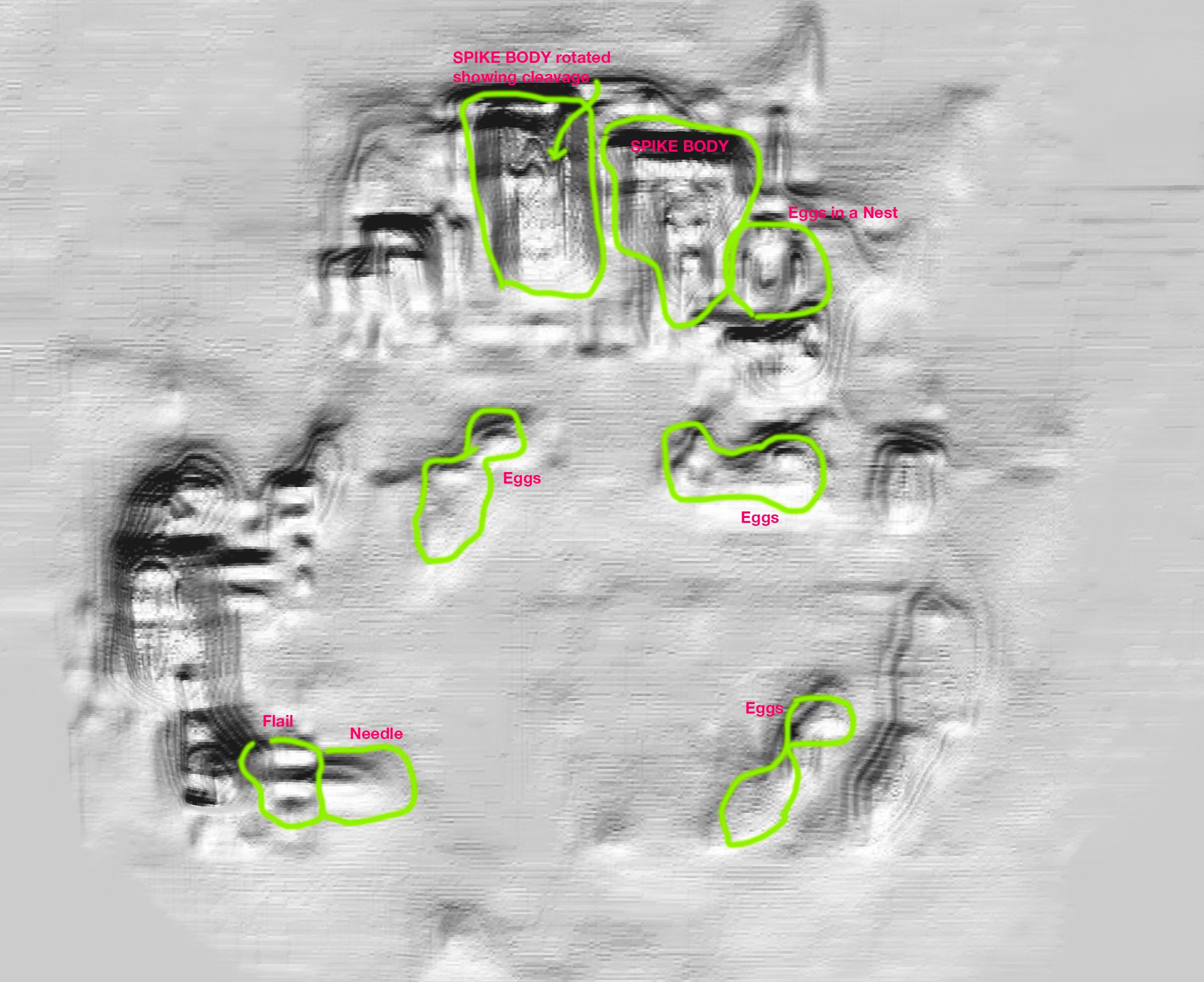Digital Holographic Visualization of SARS-CoV-2 Spike Protein
How do you study a virus doing its work—without freezing it in time?
1. SARS-CoV-2 Architecture, Illustrated from Electron Microscope Data © Molecular Architecture of the SARS-CoV-2 Virus, Hangping Yao, Et al.
Why spike architecture matters?
The SARS-CoV-2 spike protein is the key to cell entry. Traditional structural biology relies on cryo-EM: samples are cryogenically frozen, sliced into nanometer-thin sections, and reconstructed into 3D volumes(Image 1). This approach sets a gold standard for atomic-level detail—but it’s slow, costly, and fundamentally incompatible with observing dynamic behavior in natural environments.
Because we can’t actually observe these processes in a live natural environment we have to develop an understanding of the mechanics of the architectures. One of the most interesting aspects of the SARS-CoV-2 virus the site of the furin cleavage. If you will the missing bits of the spike that would normally keep the ‘petals’ at the top from opening much like the tongue on your shoe sneaker when you undo your laces. The process of propagation of the virus to the host is driven by this mechanism. So the consequence of loose laces and flapping tongues increased infection rates. As the virus spike opens and sends the precursor eggs that penetrate the host cell wall, a needle and flail architecture detach and unfold from the virus membrane move through the spike and out the top before penetrating the host cell membrane for infection (Image 2). The virus will hijack the mechanisms for replication and the virus destroys the host cell.
2. Computer Rendering of SARS-CoV-2 spike penetrating host cell
Our approach: DHOM for live, label-free super-resolution
At NanoFraction we have developed an optical super resolution method to record the SARS-CoV-2 virus in a live environment. Our approach: DHOM for live, label-free super-resolution NanoFraction’s Digital Holographic Optical Microscopy (DHOM) records interferometric holograms of specimens in liquids, gases, or solids—label-free, in situ, and in real time. We capture optical phase information that can be reconstructed to reveal sub-diffraction details and mesoscale organization—offering a complementary, triage-friendly front end to electron-microscopy pipelines.
• Live environments: Observe spike-envelope architectures without freezing or staining.
• Super-resolution phase data: Extract quantitative geometry and material contrast from the hologram.
• Fast triage: Use DHOM to prioritize samples for cryo-EM, accelerating end-to-end discovery.
The hologram recorded is seen in the image below (Image 3). We propose that NanoFraction’s DHOM technology can be an accelerator and triage technology for electron microscope pipelines. For those looking for label-free super-resolution in natural environments DHOM is a well suited approach.
Image 3 shows various architectural features that can be compared to Image 1. We have two spike proteins visible at the top of the image. To the right at the top we also see the ‘Eggs in a Nest’ architecture. On the left side of the image we see the flail and needle architecture folded into the virion membranes. Lastly we see the ‘eggs’ in the center of the membrane. The eggs contain the precursor that will contaminate the cell and will replicate the COVID-19 virus, destroying the host cell.
3. Digital Hologram of SARS-COV-2 Architecture © Nanofraction 2023
If you compare the architectures visible in the NanoFraction spike envelope recorded with the NanoFraction nanoscope with the values measured with electron microscopy the values agree. In Image 4, you see the stem inserted into the membrane of the COVID-19 virus. The shape and dimensions of the pike are derived with analytic metrology software tuned for NanoFraction nanoscope. By selecting the microscope sensitivity, it is possible to reach a molecular level resolution giving the molecular composition inside the spike envelope.
Bottom line: DHOM doesn’t replace cryo-EM—it de-risks and accelerates it by letting you see functional architectures in their native environment first, then commit the most informative samples to EM.
Where this is going?
• Correlated workflows: DHOM pre-screen → EM deep-dive → integrated model of spike mechanics.
• Kinetics in context: Track conformational readiness (e.g., post-cleavage states) under varying conditions.
• AI-assisted metrology: Automated detection and measurement of envelope features across large datasets.
4. Digital Holographic Envelope of SARS-CoV-2 Spike protein © Nanofraction 2023
Interested in collaborating?
If you’re building correlated optical–EM pipelines or exploring label-free super-resolution in native environments, we’d love to compare notes and data. Contact us to discuss a joint study or pilot.